|
I am a research scientist at Genentech, Prescient Design team. Previously, I was a research scientist at Deep Genomics and Element AI (where I work closely with people from Mila). I received a Ph.D. from École Polytechnique Fédérale de Lausanne (EPFL) and Idiap Research Institute (Switzerland), under supervision of Ronan Collobert. During my phd, I also spent time at Facebook AI Research (FAIR). Previously, I graduated with a MSc. in Image and Signal Processing from Institut National des Sciences Appliquées de Lyon (INSA), in France. I am originally form sunny Fortaleza, Brazil. |

|
|
I am interested in machine learning and its applications, specifically deep learning methods for representation learning and generative modeling.
Currently, I am interested in developing AI methods that can tackle complex scientific problems, such as molecular sciences and drug discovery.
|
|
|
Pedro O. Pinheiro, Pan Kessel, Aya A. Ismail, Sai P. Mahajan, Kyunghyun Cho, Saeed Saremi, Nataša Tagasovska NeurIPS, 2025 paper | code (coming soon) |
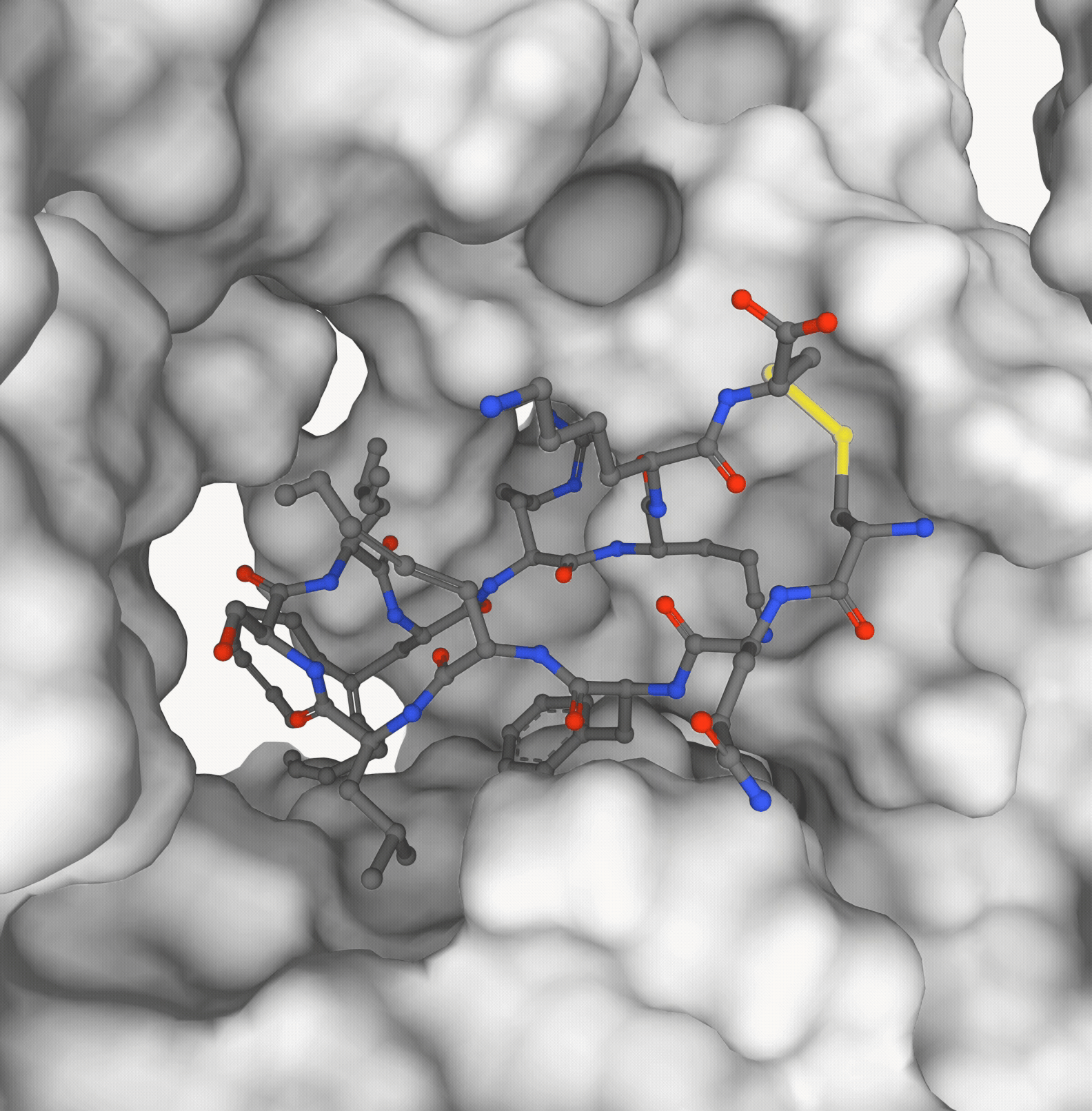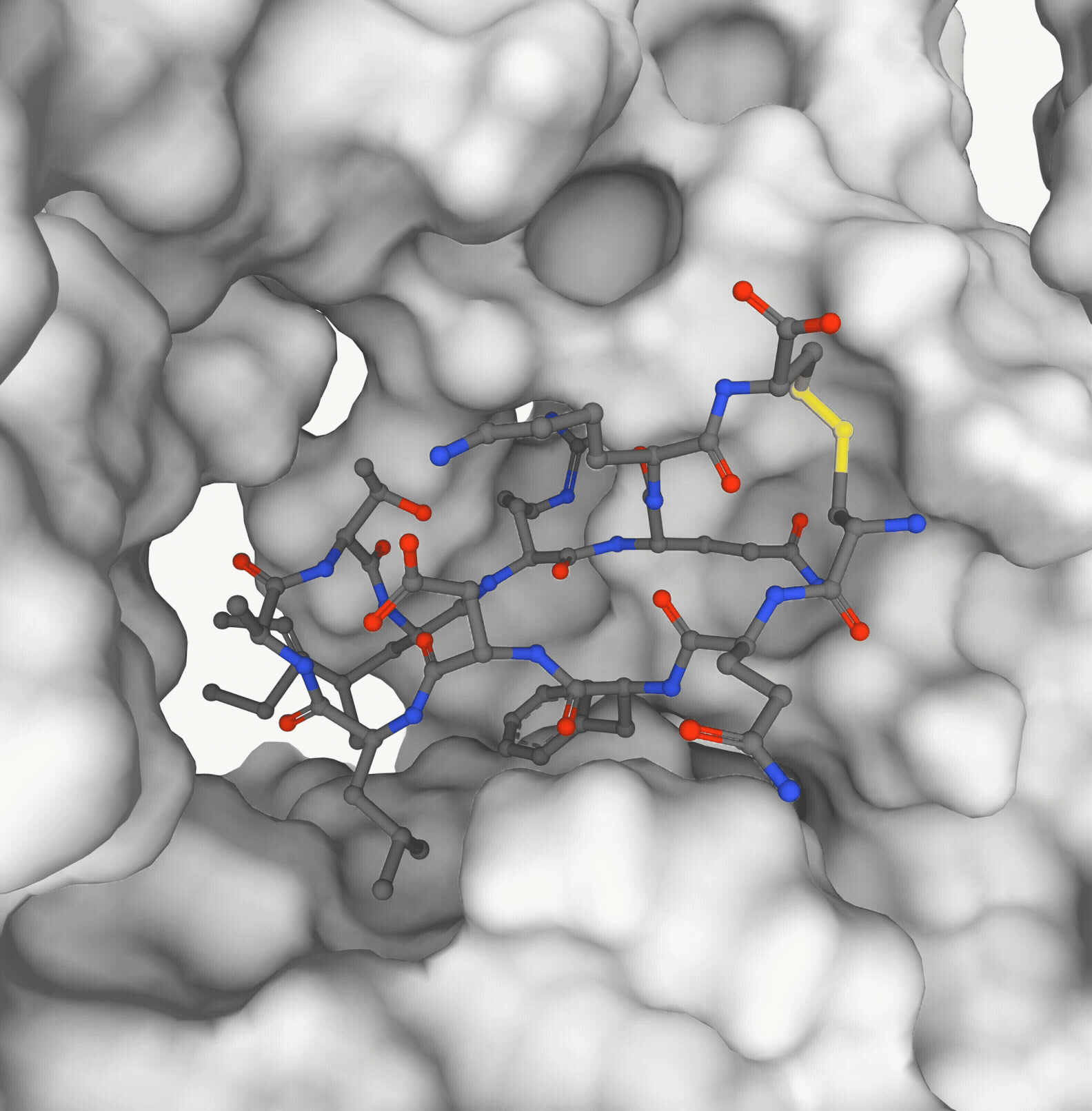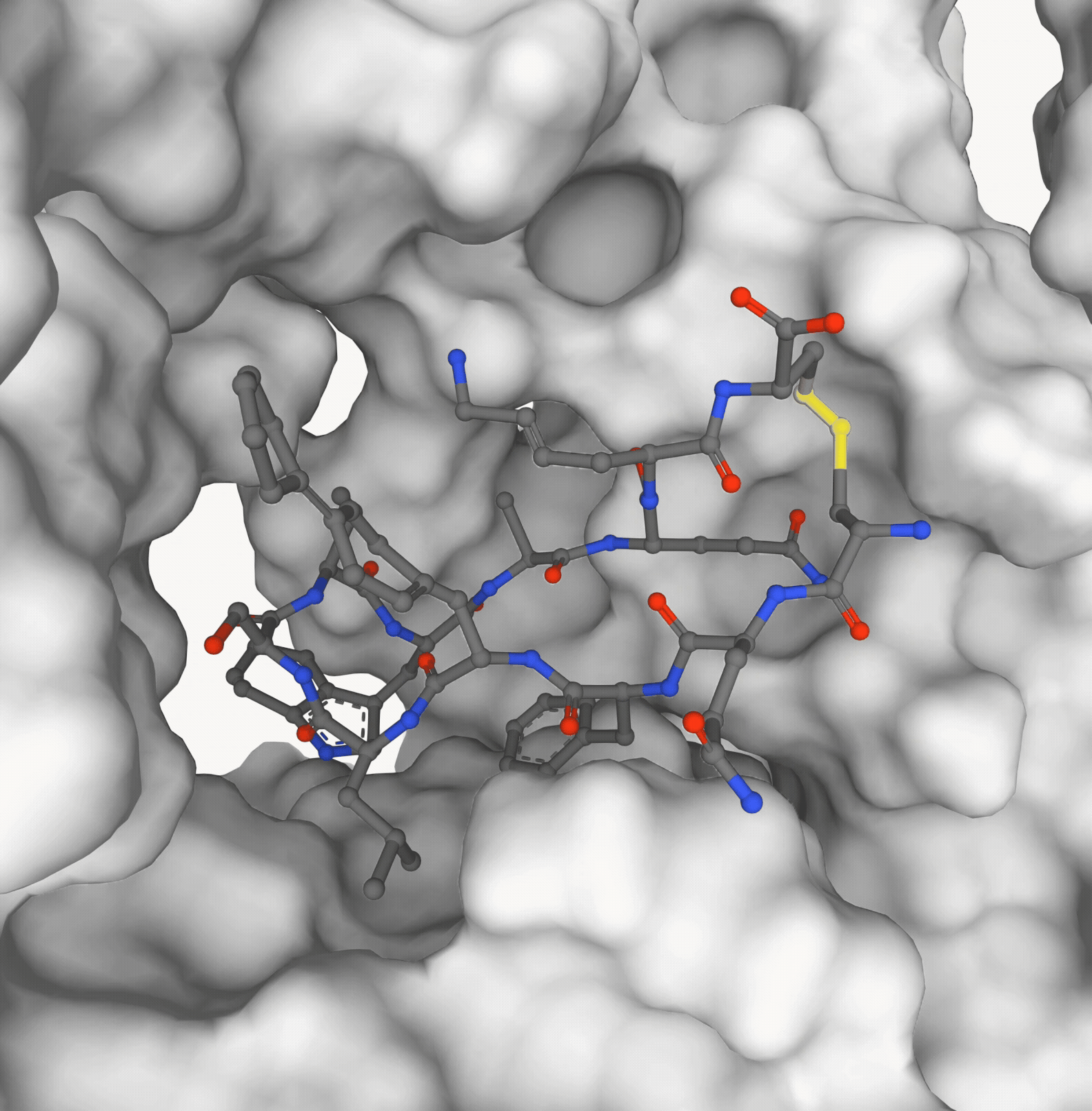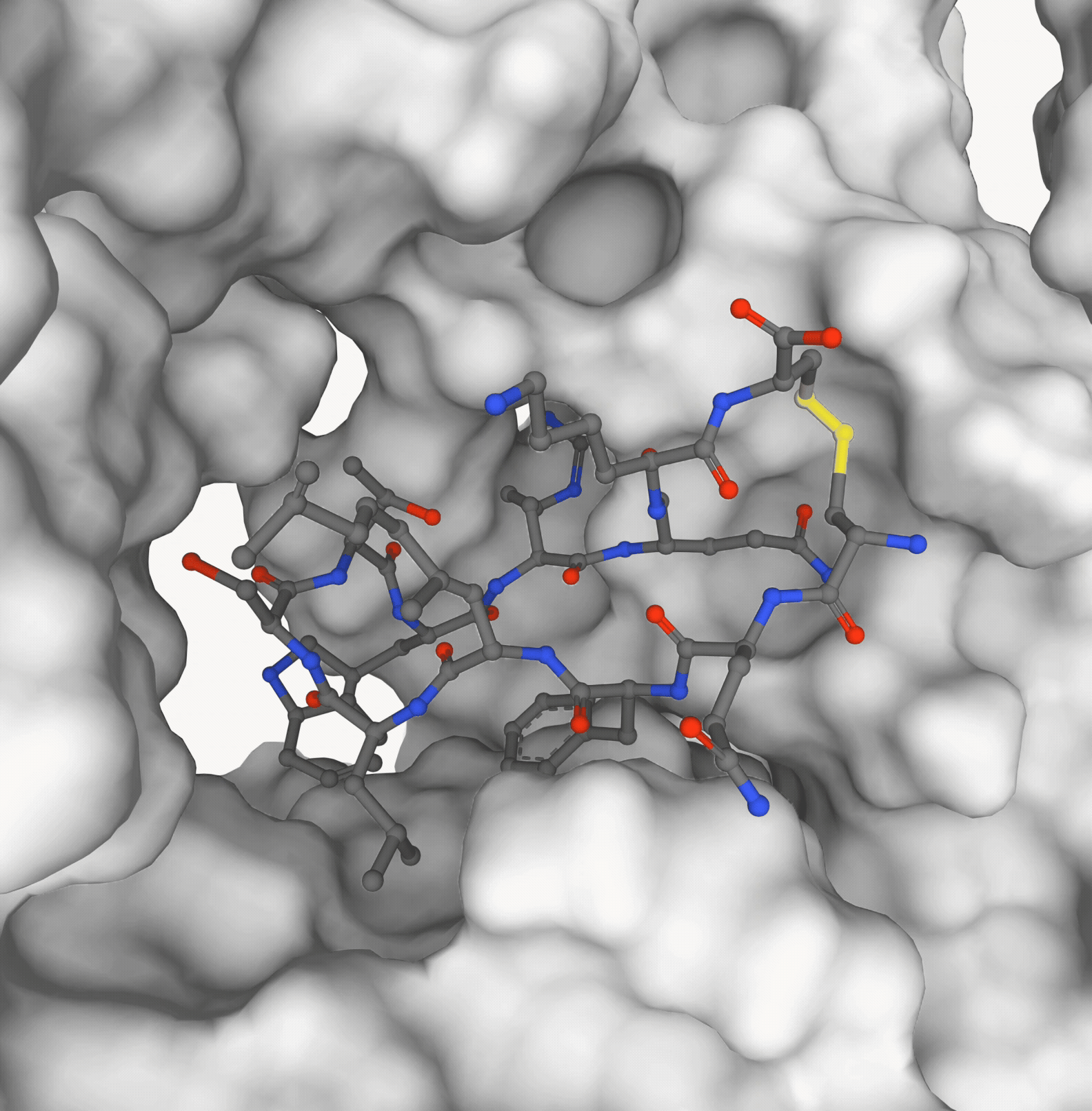
|
Matthieu Kirchmeyer*, Pedro O. Pinheiro*, Karolis Martinkus, Emma Willett, ..., Richard Bonneau, Saeed Saremi NeurIPS, 2025 (*equal contribution) paper | code (coming soon) |

|
Matthieu Kirchmeyer*, Pedro O. Pinheiro*, Saeed Saremi NeurIPS, 2024 (*equal contribution) paper | code |

|
Pedro O. Pinheiro, Arian Jamasb, Omar Mahmood, Vishnu Sresht, Saeed Saremi ICML, 2024 paper | code |

|
Pedro O. Pinheiro, Joshua Rackers, Joseph Kleinhenz, Michael Maser, Omar Mahmood, Andrew Martin Watkins, Stephen Ra, Vishnu Sresht, Saeed Saremi NeurIPS, 2023 paper | code |

|
Joshua Yao-Yu Lin, Jennifer L. Hofmann, Andrew Leaver-Fay,..., Pedro O. Pinheiro,..., Andrew Watkins, Kyunghyun Cho, and Nathan C. Frey bioRxiv, 2025 paper |
|
Albi Celaj, Alice Jiexin Gao, Tammy T.Y. Lau, Erle M. Holgersen, ..., Pedro O. Pinheiro, ..., Brendan J. Frey bioRxiv, 2023 paper |